Research News
Comparison and optimization of CRISPR/dCas9/gRNA genome-labeling systems for live cell imaging
The dynamic localization of a particular genomic locus in a three-dimensional (3D) genome has been proposed to regulate various genome functions including gene transcription, DNA recombination, DNA replication, and DNA repair. Until recently, several strategies have been developed to trace the dynamic movement of genomic loci in living cells. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9), an RNA-guided endonuclease that mediates highly sequence-specific binding and efficient cleavage on genomic DNA, has been extensively developed recently for genome editing. On the other hand, a nuclease-deficient Cas9 (dCas9) could bind to a guide RNA (gRNA)-specific genomic locus, where by recruiting various effectors it could achieve precise and programmed transcription activation and repression, epigenetic remodulations of local histone and DNA modifications, labeling and visualization of the genomic locus, and single base genome mutagenesis. Various dCas9/gRNA systems have been designed to label genomic loci in live cells. In its first version, direct fusion of fluorescent proteins such as green fluorescent protein (GFP) with dCas9 protein was used by Huang’s laboratory. To increase the signals, a SunTag that contains multiple copies (24X) of GCN4 peptide epitopes has been added to the C-terminal dCas9. Fusion with single-chain fragment variable (scFv) antibody against GCN4 peptide allows more copies of fluorescent proteins to be recruited to a single tethered dCas9/gRNA complex. Recently, tandem FP11-tags were also fused to dCas9 to allow proportional enhancement of the fluorescence signal. To achieve simultaneous labeling of several genomic loci at the same time, two approaches have been developed. First, several CRISPR/Cas9 orthologous proteins from distinct bacterial species that have different gRNA-binding specificities could be fused to different fluorescent proteins. On the other hand, both RNA aptamer binding effectors and Pumilio/FBF (PUF) RNA-binding proteins have been utilized to label the different gRNAs, which could work with the same dCas9 protein. In addition, multiple copies of RNA motifs could be fused to the gRNA to greatly amplify the signals.
In this study, researchers compared several of the latest gRNA labeling and dCas9 labeling systems in the same experimental settings such as the cell type, transfection method, and gRNA expression cassette, as well as genomic targets. They have identified and solved the intrinsic nonspecific labeling issue associated with the gRNA labeling methods.
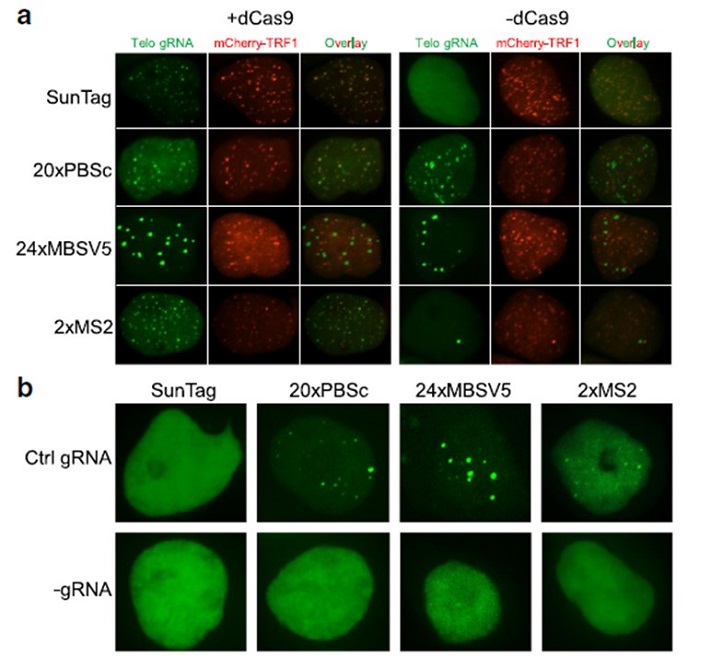
Comparison of different dCas9/gRNA systems for labeling human telomeres.
They also developed novel bimolecular fluorescence complementation (BIFC)-dCas9/gRNA methods that combine the specificities of both dCas9 and gRNAs. The BIFC-dCas9/gRNA methods have high signalto-noise ratio and no nonspecific foci.
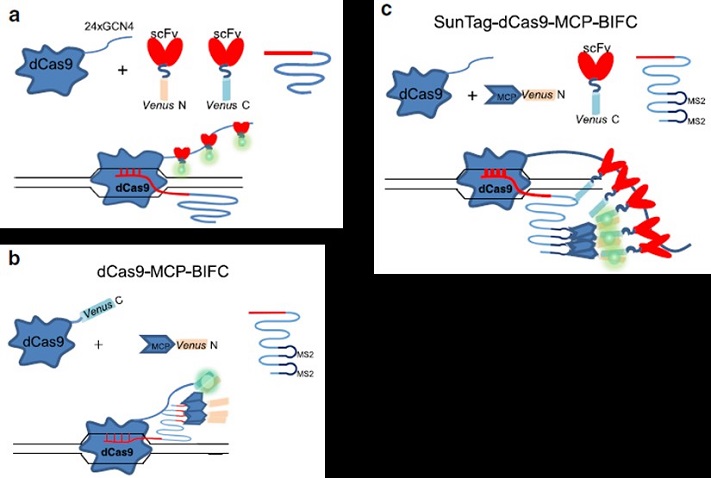
Schematic views of the different BIFC-dCas9/gRNA genome labeling systems.
Yu Hong, PTN program PhD student in Dr. Yu Zhang’s lab, is the first author of this paper. Dr. Yu Zhang is the corresponding author. Other contributing authors include Guangqing Lu, Jinzhi Duan and Wenjing Liu from Dr. Yu Zhang’s lab. This research was supported by the “Hundred, Thousand and Ten Thousand Talent Project” by the Beijing municipal government (2017A02), the “National Thousand Young Talents Program” of China, and the National Natural Science Foundation of China (81572795 and 81773304) to Y.Z.



