Institutional News
On November 8, 2018, Dr. Yu Zhang’s laboratory published a paper titled " Live imaging and tracking of genome regions in CRISPR/dCas9 knock-in mice " in Genome Biology. (https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1530-1)
Recently developed revolutionary CRISPR/Cas9 technique is becoming one of the hottest tools in biological studies for almost all model organisms. The central advantage of CRISPR/Cas9 system is its extraordinary ability to specifically and robustly recognize and cleave targeted genomic sequences in vivo, which relies on Watson-Crick base pairing between guide RNA (gRNA) and its recognition sequences. While extensive efforts have been focused on the optimization and implication of targeting and cleavage by CRISPR/Cas9 systems for genome editing, the nuclease-deactivated Cas9 (dCas9) could also be used as a versatile tool to genetically and epigenetically modulate the targeted genomic locus, and label the genomic loci in living cells. For example, dCas9/gRNA complexes could recruit transcription activators and repressors to gRNA-targeted genes and lead to gene transcription activation and repression, respectively. Fusion with other effectors containing epigenetic remodeling activities could also lead to targeted epigenetic changes such as DNA methylation/demethylation, histone modifications, as well as chromatin remodeling. Moreover, fusion with DNA deaminases could lead to precise nucleotide mutations on targeted genomic loci. Finally, various dCas9/gRNA-based strategies have been developed for live tracking the movement of chromatin regions by recruiting fluorescent proteins to the targeted genome regions.
So far, the dCas9/gRNA tools have been mostly developed in cell culture systems where the dCas9, gRNA, and effector expression cassettes could be transfected or infected into the cells. However, for in vivo applications of dCas9/gRNA in live animals, how to efficiently deliver all these components, especially the large dCas9 expression cassettes, into the cells of various tissues remains to be a major difficulty. To solve this problem, the researchers from Zhang Yu lab generated mouse strain in which dCas9-EGFP was ubiquitously expressed (Figure 1). With such unique animal models, they also investigated the role of TRF1, a shelterin protein, in regulating telomere dynamics in live animals. They developed CRISPRimaging-interference (CRISPRii) method to simultaneously repress TRF1 and label telomeres (Figure 2).
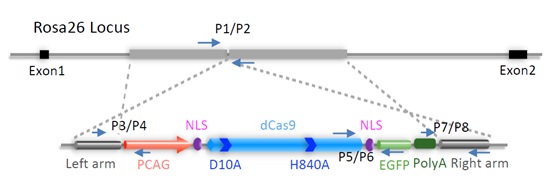
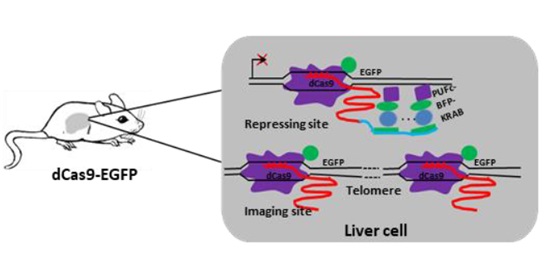
Figure 2. The schematic of CRISPR imaging and interference (CRISPRii) strategy in dCas9-EGFP mouse.
More importantly, with such dCas9-EGFP knockin mice, the researchers studied the dynamics of telomeres in live animals for the first time. Indeed, they observed surprising results which are very different from that obtained in cultured cell lines (Figure 3),urging the necessity of studies in vivo. This research applied dCas9/gRNA labelling system into live animals for the first time and it provides an important tool for the field to study chromatin 3D structure and dynamics.
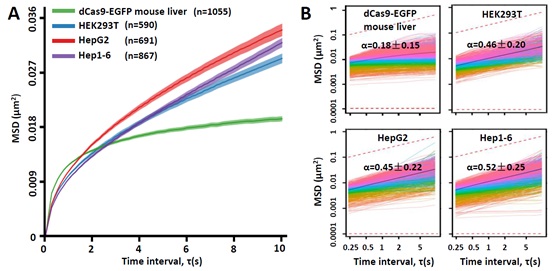
Figure 3. Unique features of telomere dynamics in mouse liver revealed by dCas9-EGFP knock-in mice
Jinzhi Duan, Guangqing Lu, Yu Hong and Qingtao Hu in Dr. Yu Zhang’s lab, are co-first authors of this paper. Dr. Yu Zhang is the corresponding author. Other contributing authors include Xueying Mai, Jing Guo, Xiaofang Si from Dr. Yu Zhang’s lab and Dr. Fengchao Wang from Transgenic Animal Center in NIBS. This research was supported by National Natural Science Foundation of China (81572795 and 81773304), the “Hundred, Thousand and Ten Thousand Talent Project” by Beijing municipal government (2017A02). Q.H. is support by National Natural Science Foundation of China (31701135).



