Research News
isHCR: A hybridization-chain-reaction-based method for amplifying immunosignals
On Feburary 26th ,2018, Dr. Minmin Luo’s laboratory published a paper entitled “A hybridization-chain-reaction-based method for amplifying immunosignals” in Nature Methods. In this study, they described the development of immunosignal HCR (isHCR), a new signal amplification system for detecting biomolecules.
Antibody-based immunoassays remain the most popular methods for detecting and identifying the location of proteins and other biomolecules in biological samples. A major limitation of immunoassays is that the low abundance of a given target molecule in a sample often necessitates signal amplification to make detection possible. Amplification can be achieved with conjugated enzymes, but current amplification methods have several drawbacks such as high background, reduced spatial resolution, and lack of multiplexity; in addition, they are not suitable for use with large-volume samples in newer tissue-expansion and clearing techniques.
To overcome many of these limitations, researchers from Dr. Minmin Luo’s lab developed isHCR. isHCR adopts hybridization chain reaction (HCR) technology for immunosignal amplification. HCR is an enzyme-independent reaction based on recognition and hybridization events that occur between sets of DNA hairpin oligomers that self-assemble into polymers. Researchers conjugated the HCR initiators to antibodies therefore incorporated HCR reaction into traditional IHC as an amplification step. They successfully applied isHCR to enhance immunohistochemistry (IHC) staining signals in western blotting, cultured cells, and tissues.
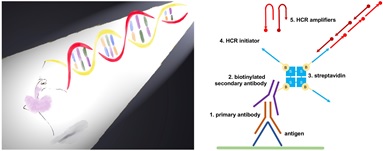
Schematic of isHCR
Researchers further optimized isHCR by using graphene oxide (GO) to suppress the background noise. They also developed a multi-round amplification method, named isHCRn, based on the basic isHCR system. They demonstrated that isHCR allows sensitive detection of many proteins in mouse brain samples and the in vivo translocation of bacterial effectors.

Sensitive detection of Vglut3 in mouse brain sections by optimized isHCR
Furthermore, they directly conjugated DNA HCR initiators to secondary antibodies via chemical linkers. This modification allows simultaneous isHCR amplification of multiple targets. They successfully performed multiplexed isHCR amplification using initiator-labeled secondary antibodies in western blotting, immunostaining of cultured cells, and immunostaining of brain sections. They also demonstrated that a variety of genetically-engineered protein tags could be added to target proteins in cells to enable the direct binding of targets to HCR initiators. These extensions of the isHCR concept beyond biotin-streptavidin interactions and beyond primary and secondary antibodies demonstrate that isHCR can be used effectively for highly multiplexed signal detection.
Recent developments in tissue preparation techniques are revolutionizing the high-resolution imaging of large-volume samples. Researchers also optimized isHCR for the use in expansion microscopy and uDISCO tissue clearing.

Optimization of isHCR for multiplexed labeling,
expansion microscopy (ExM) and uDISCO clearing
Considering the vast expanse of biotechnologies (e.g., protein tags, DNA modifications, reaction chemistries, etc.) that can potentially be incorporated into isHCR, it is conceivable that a great many biosensors and antibody-based methods in life science research could be improved or extended via the creative application of these methods.
Rui Lin, PTN program PhD student in Dr. Minmin Luo’s lab, is the first author of this paper. Dr. Minmin Luo is the corresponding author. Other contributing authors include Qiru Feng and Ruiyu Wang from Dr. Minmin Luo’s lab; Dr. Feng Shao, Dr. Peng Li and Dr. Ping Zhou from Dr. Feng Shao’s lab in NIBS; Dr. Nan Tang and Dr. Zhe Liu from Dr. Nan Tang’s lab in NIBS; Dr. Xiangbing Qi and Zhiqiang Wang from Chemistry Center in NIBS. This study was supported by MOST 973 grants, NNSFC, and Beijing Municipal Government. All work was completed in NIBS.



